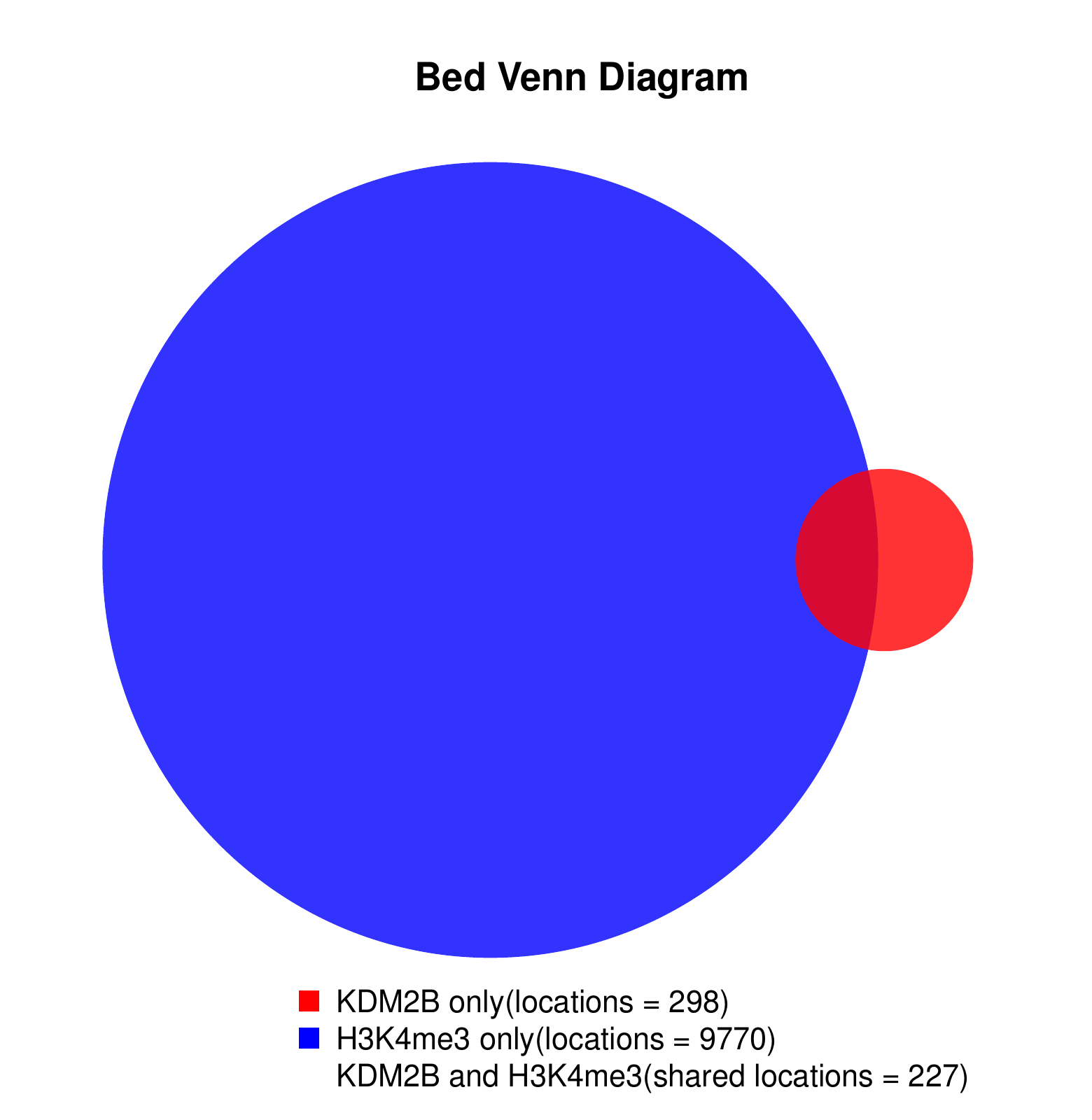
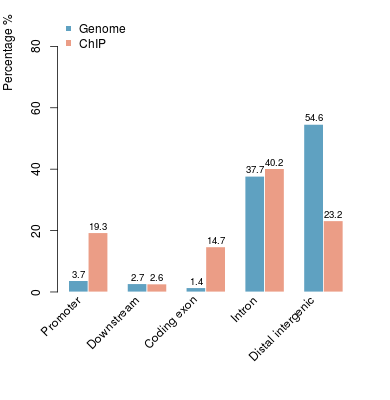
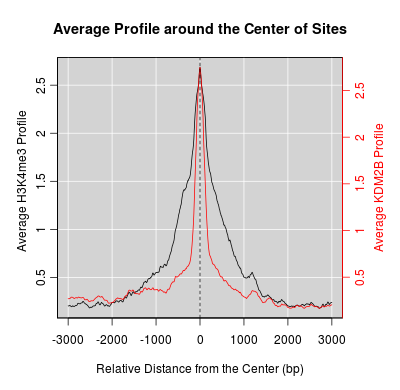
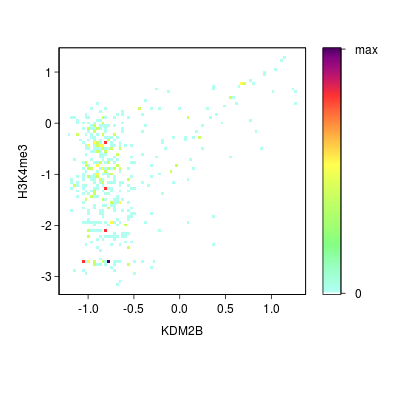
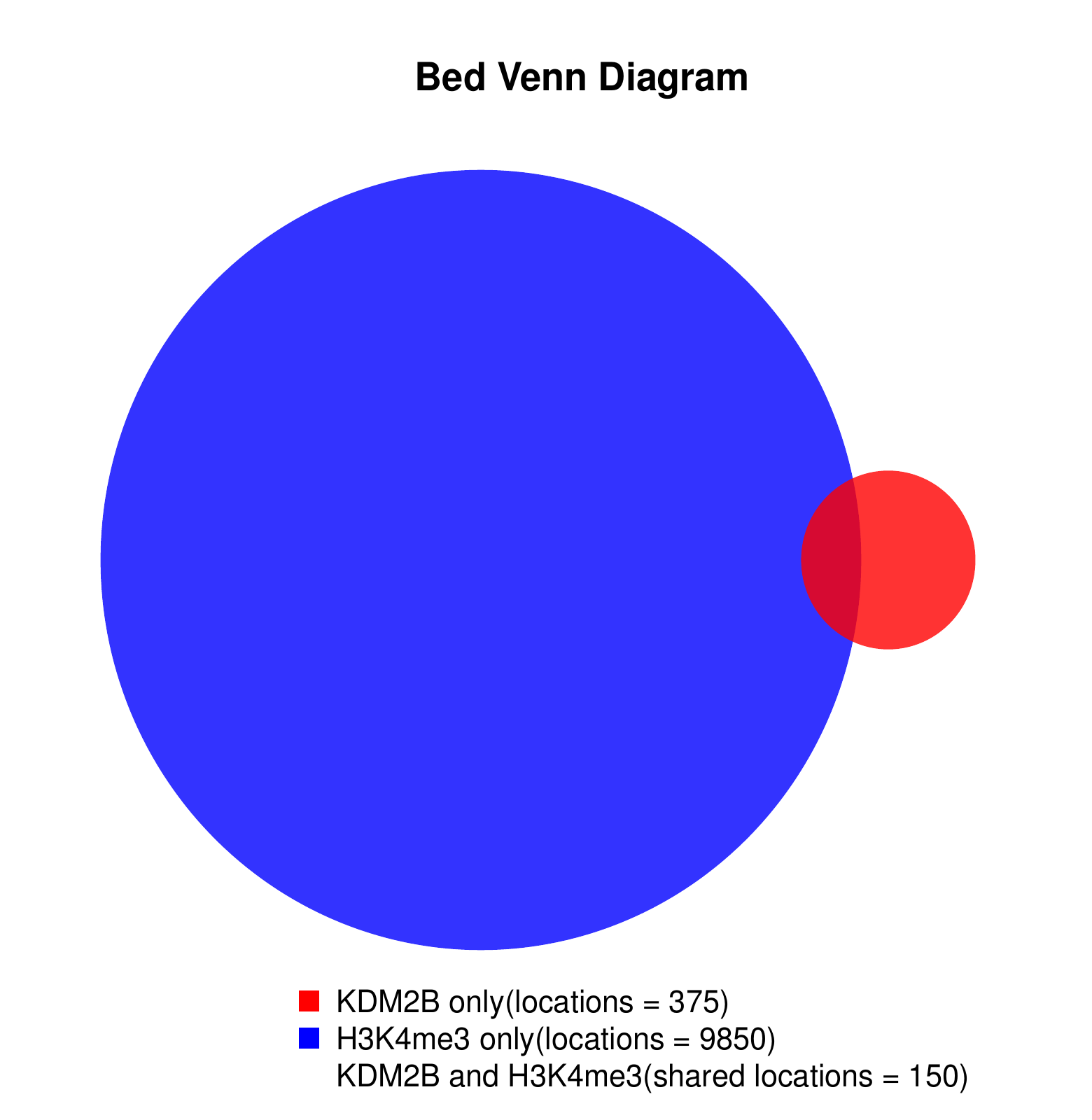
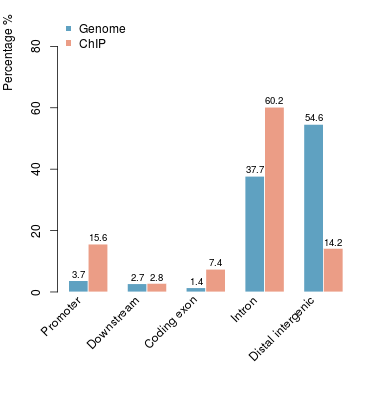
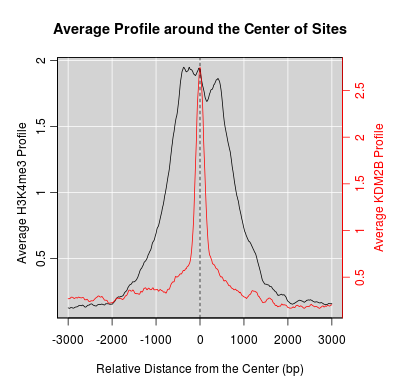
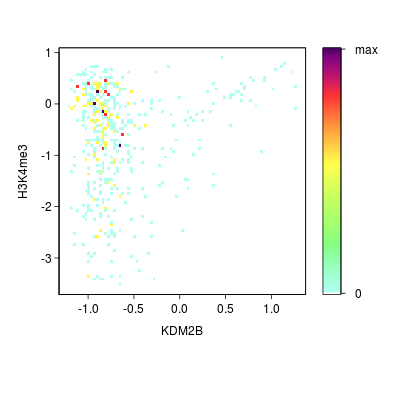
Go Back
Go Back
Comparison: |
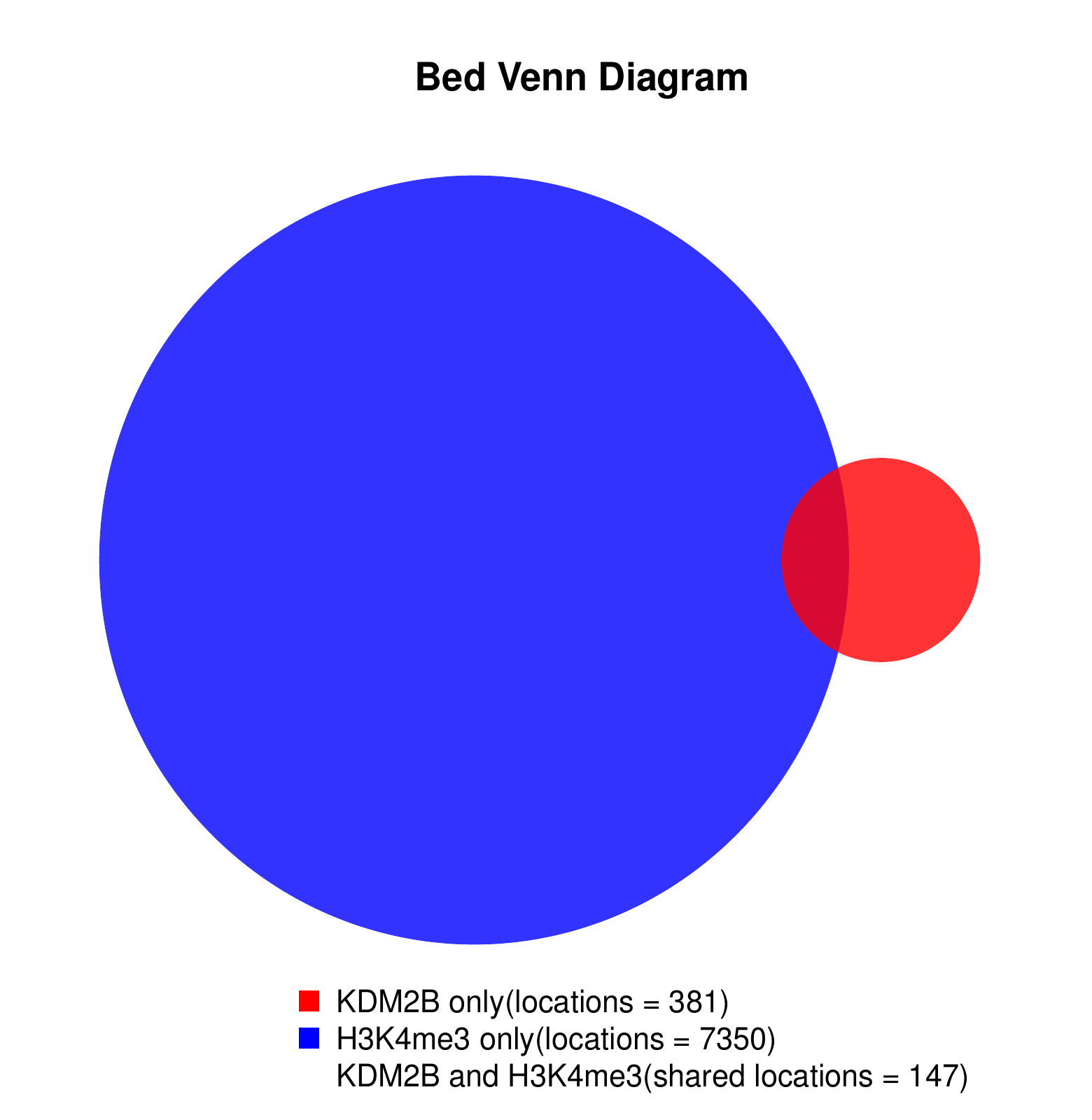
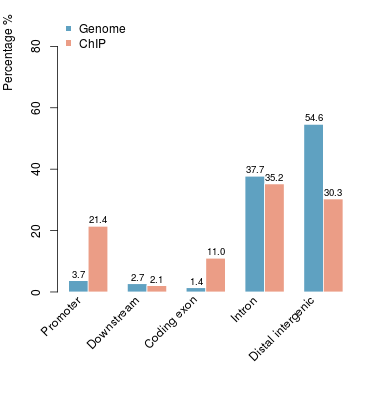
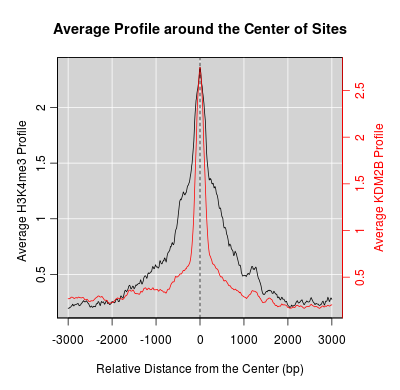
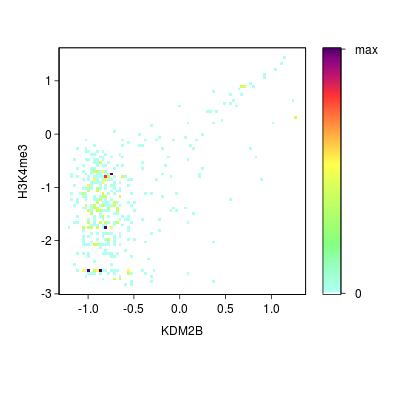
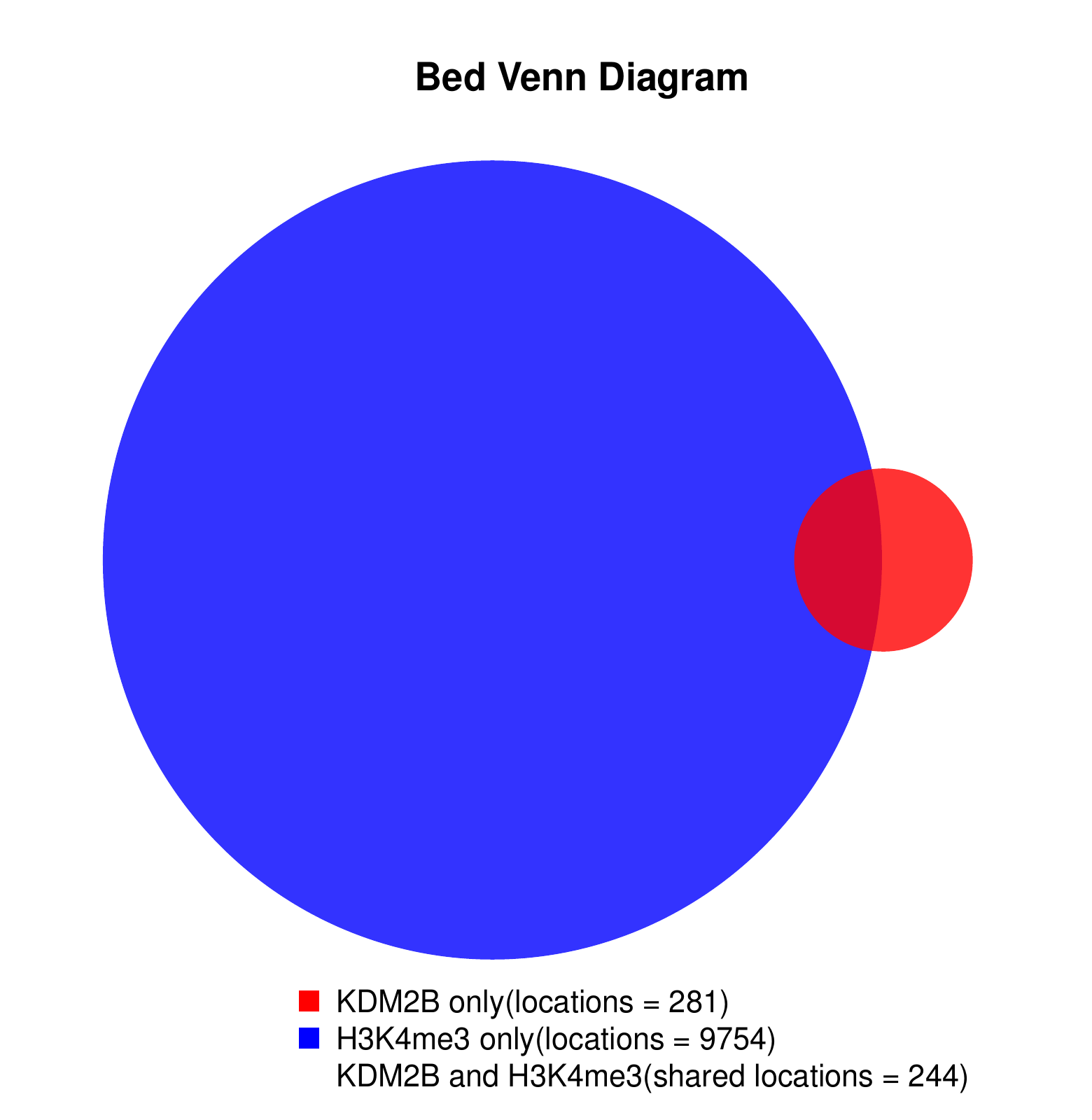
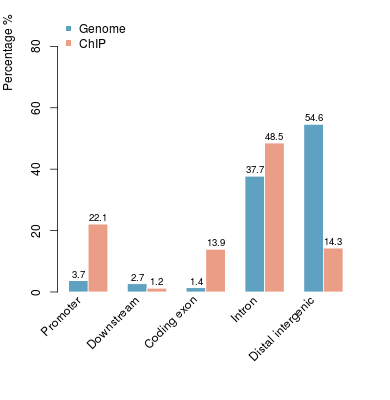
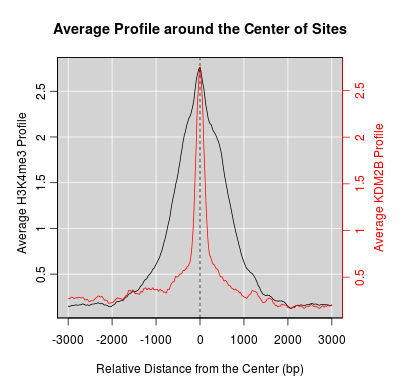
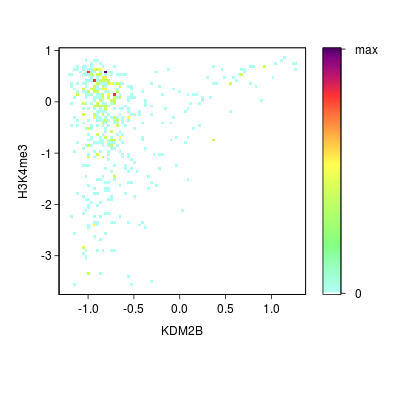
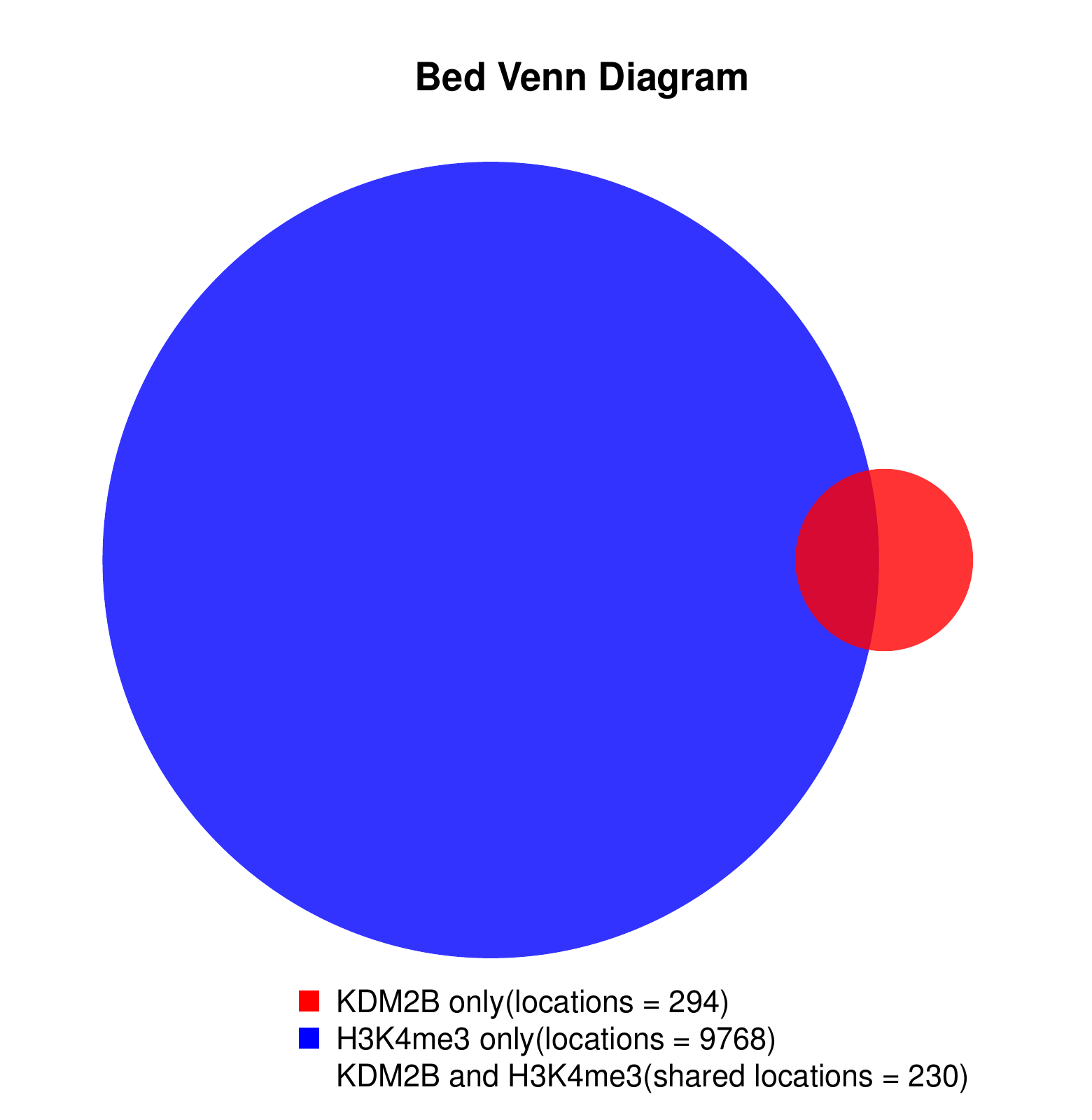
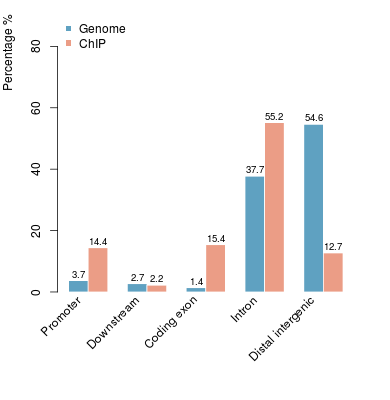
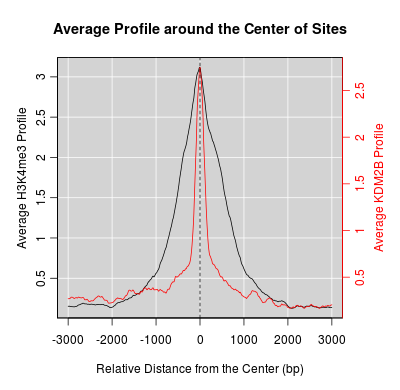
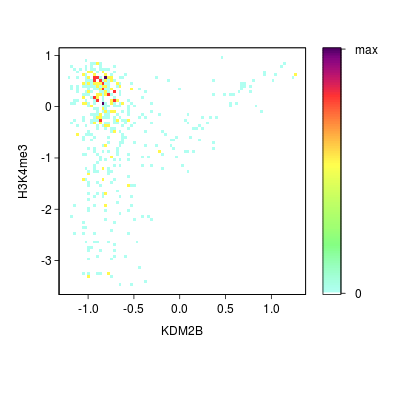
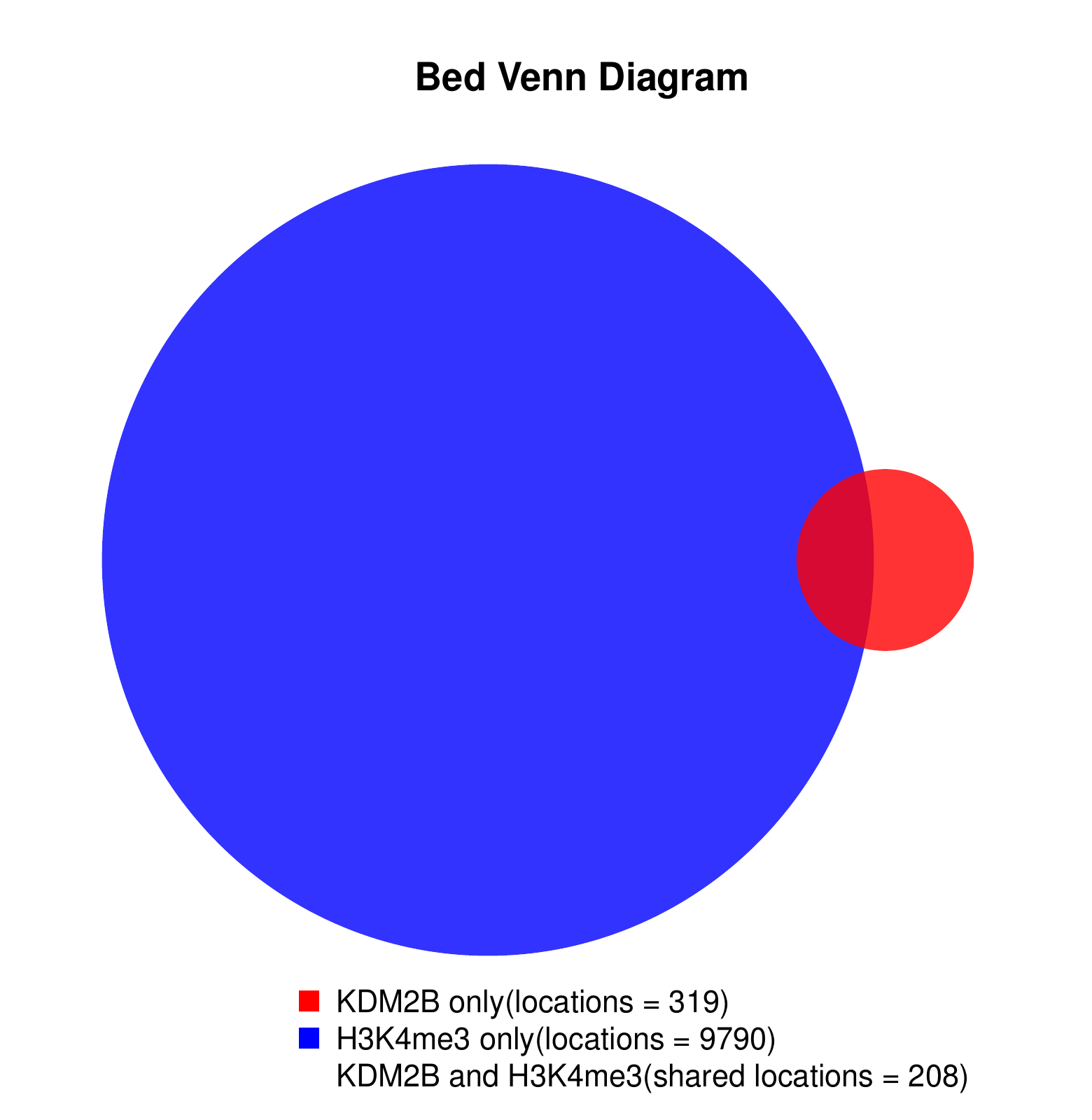
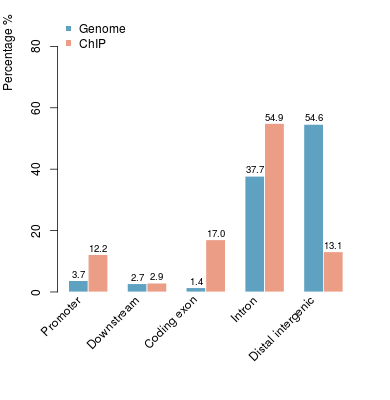
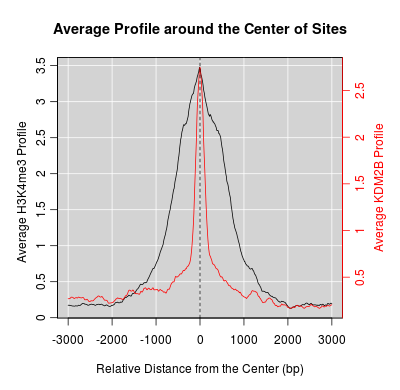
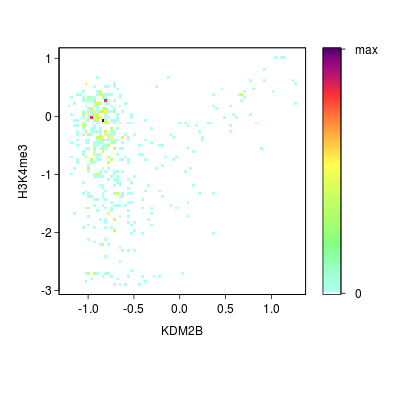
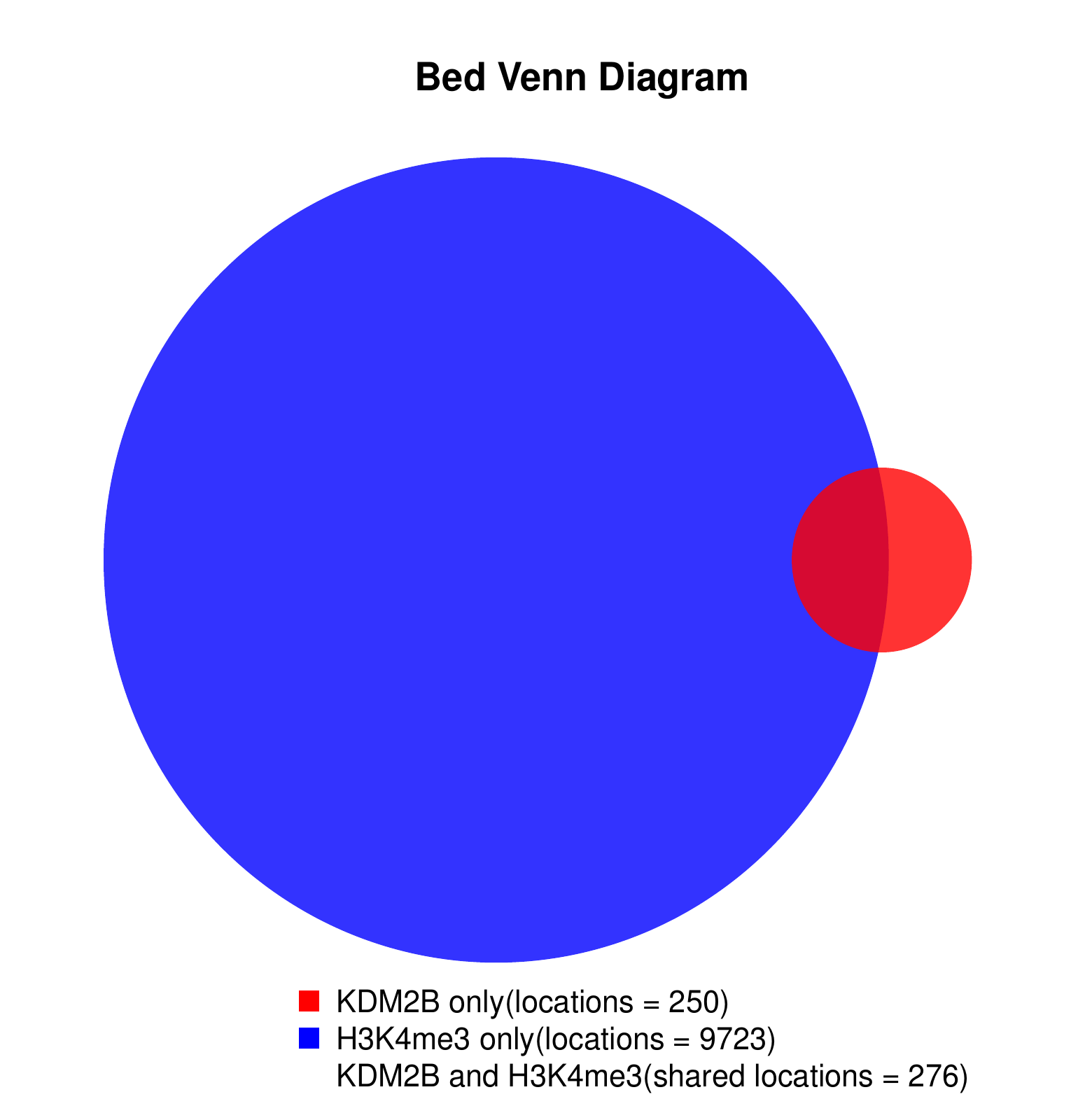
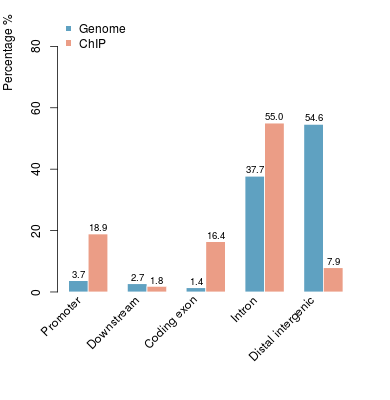
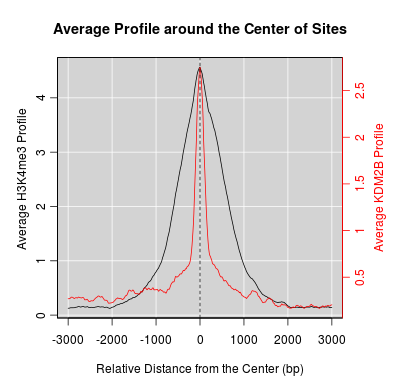
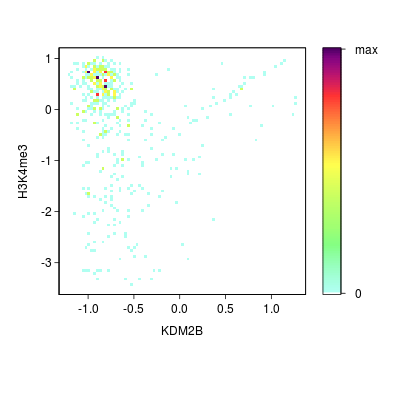
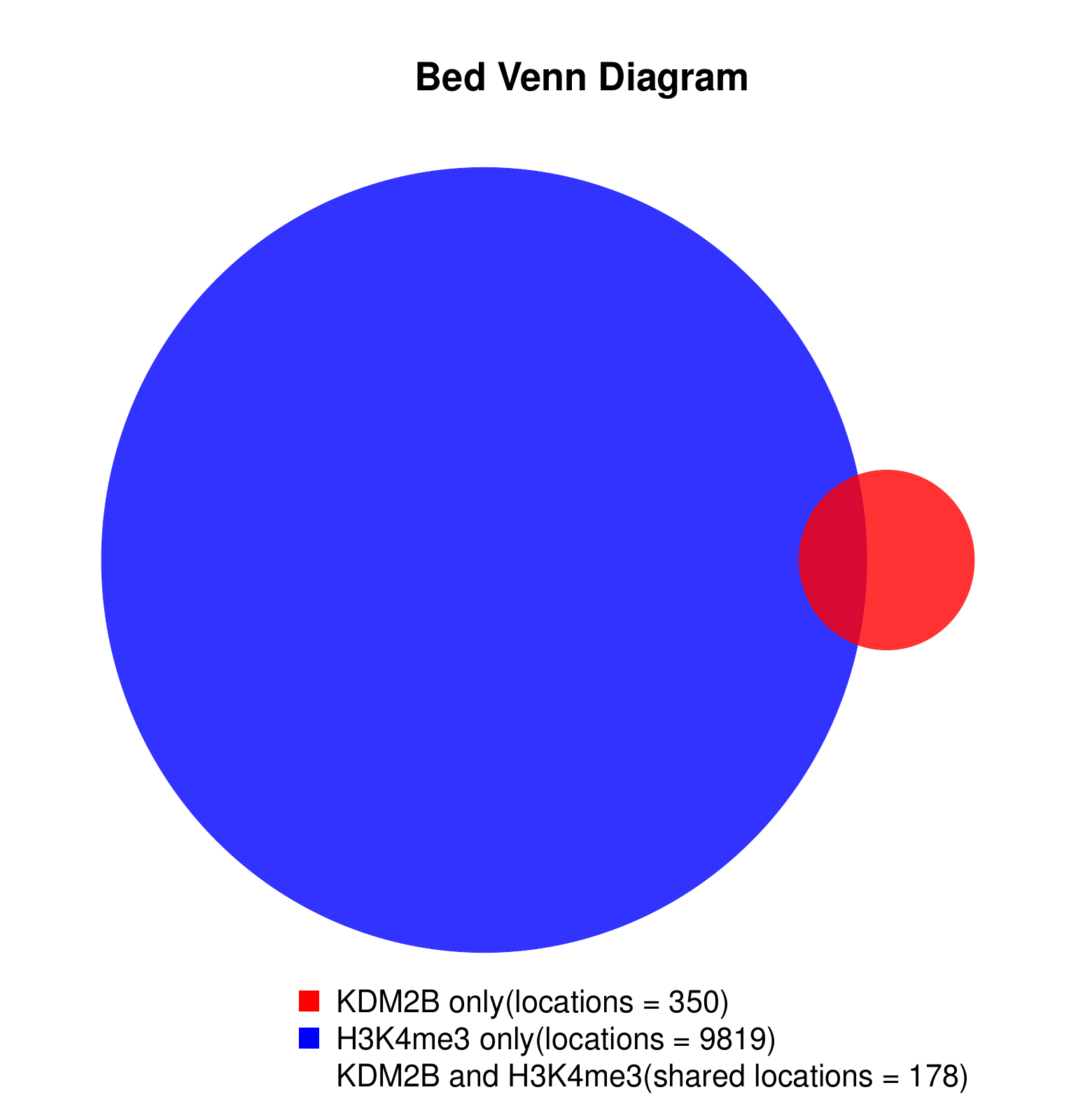
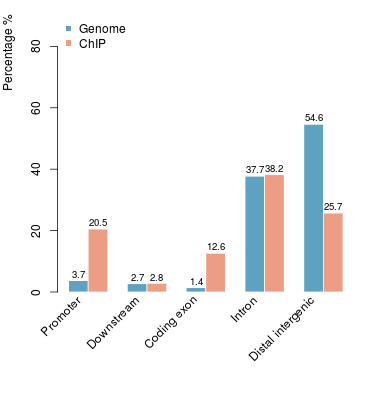
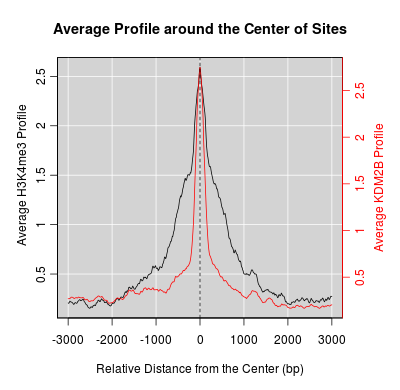
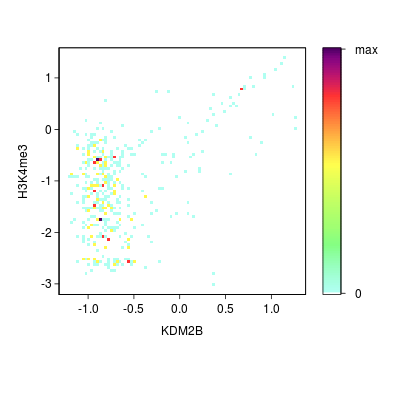
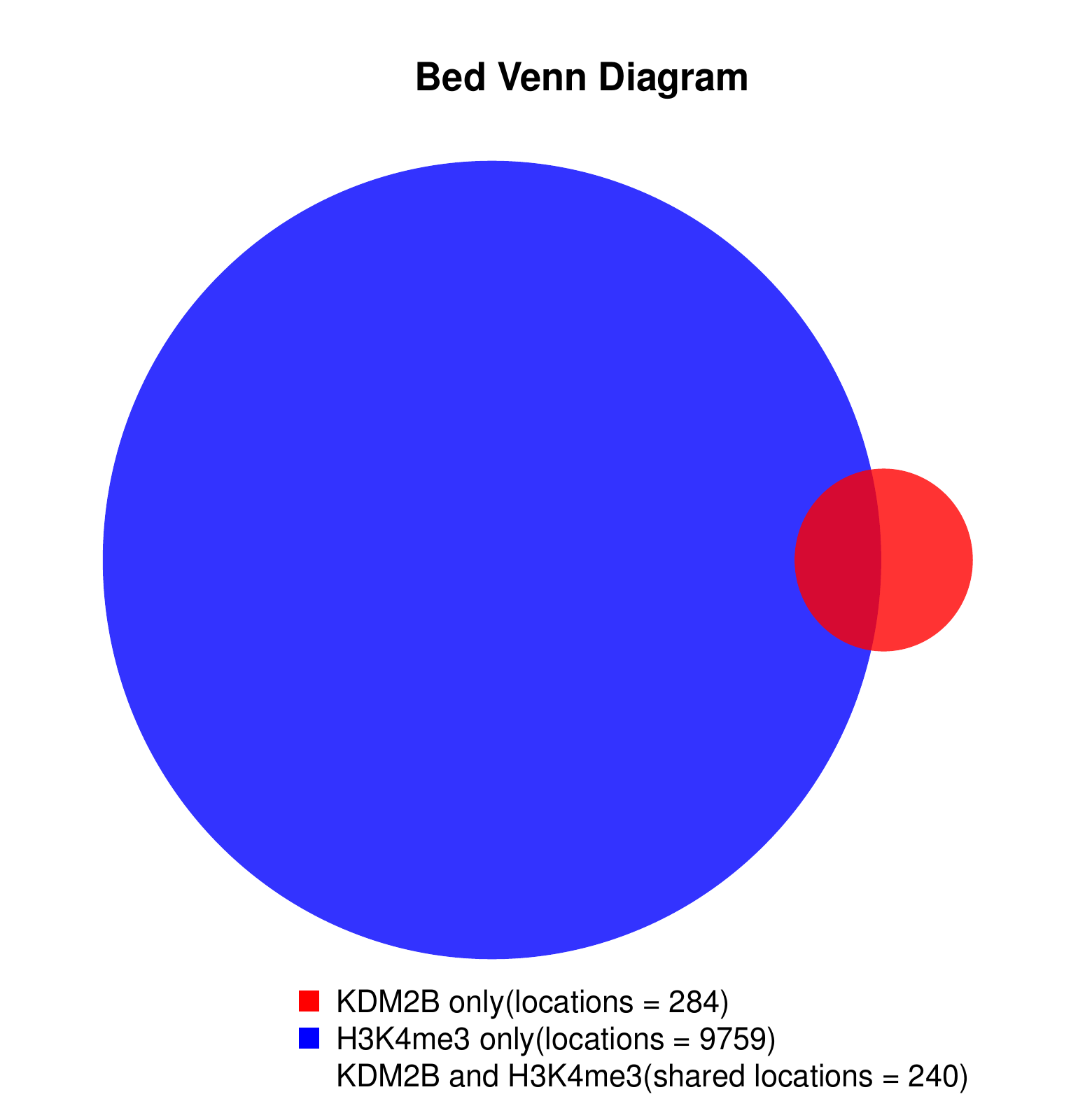
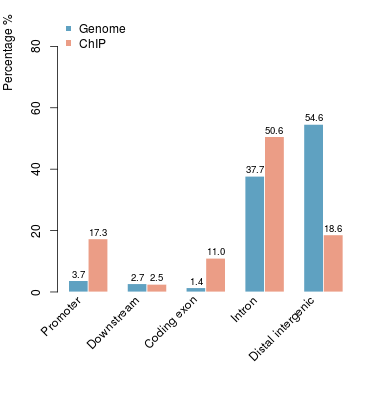
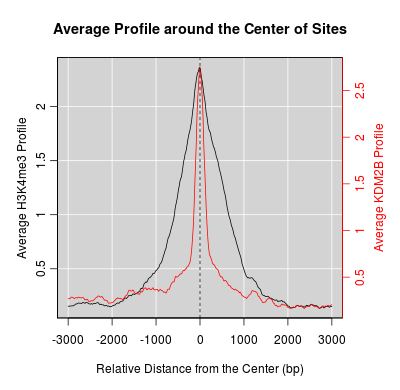
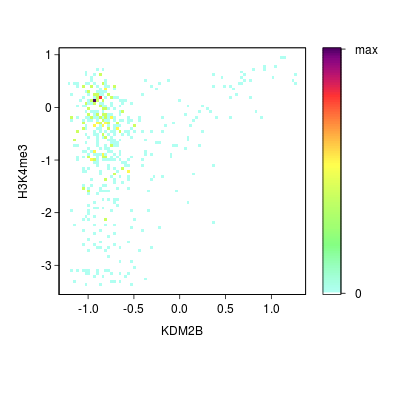
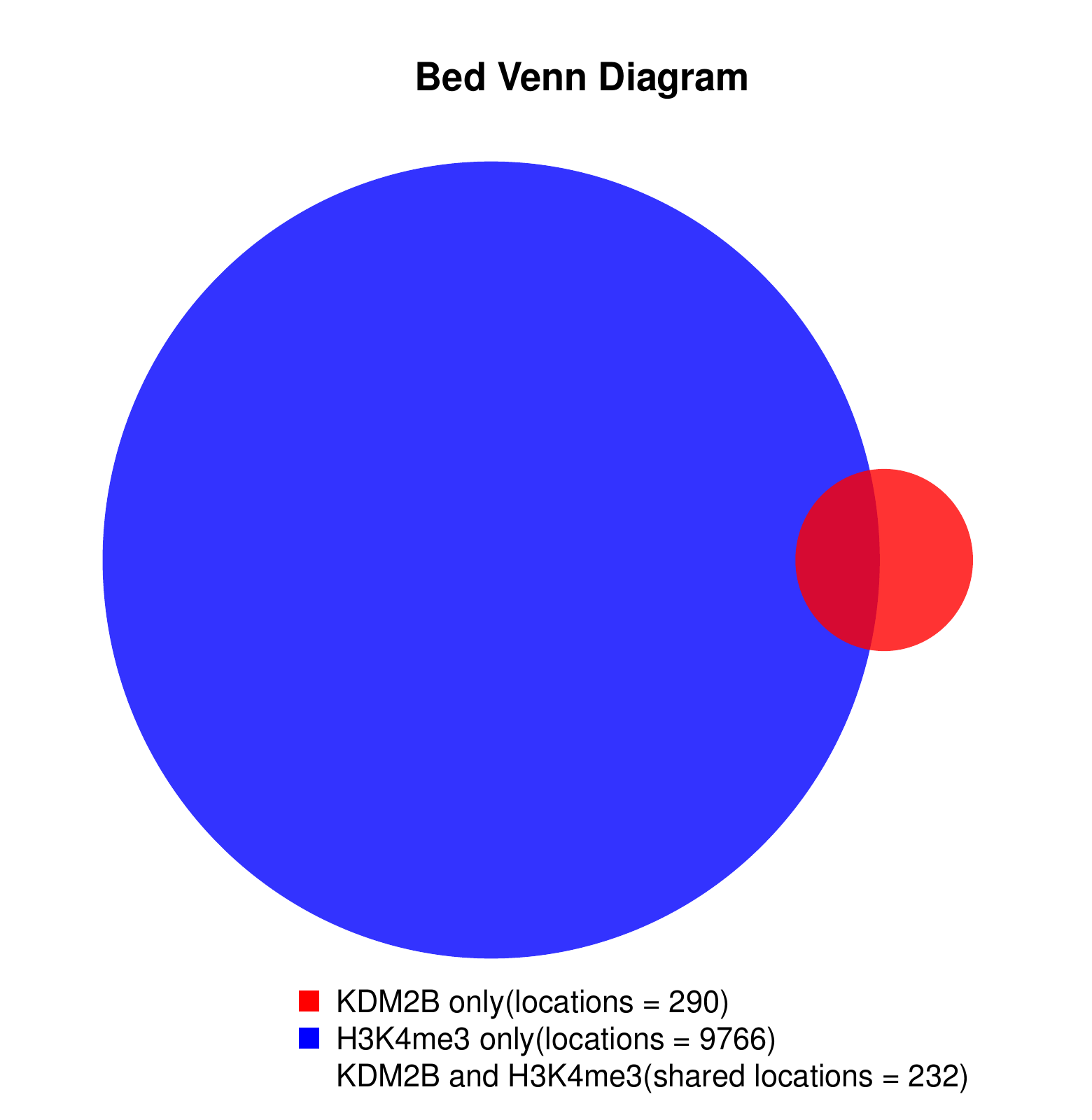
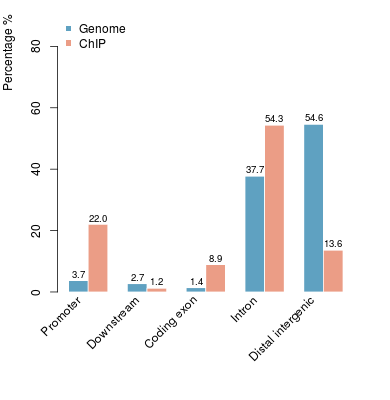
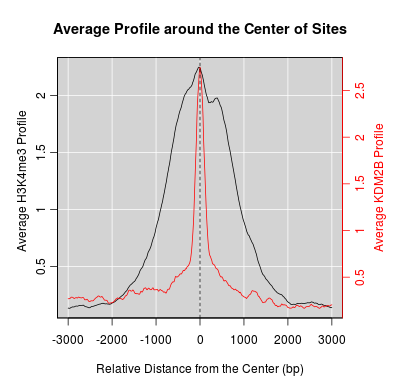
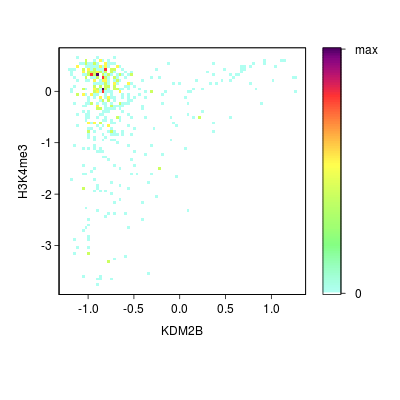
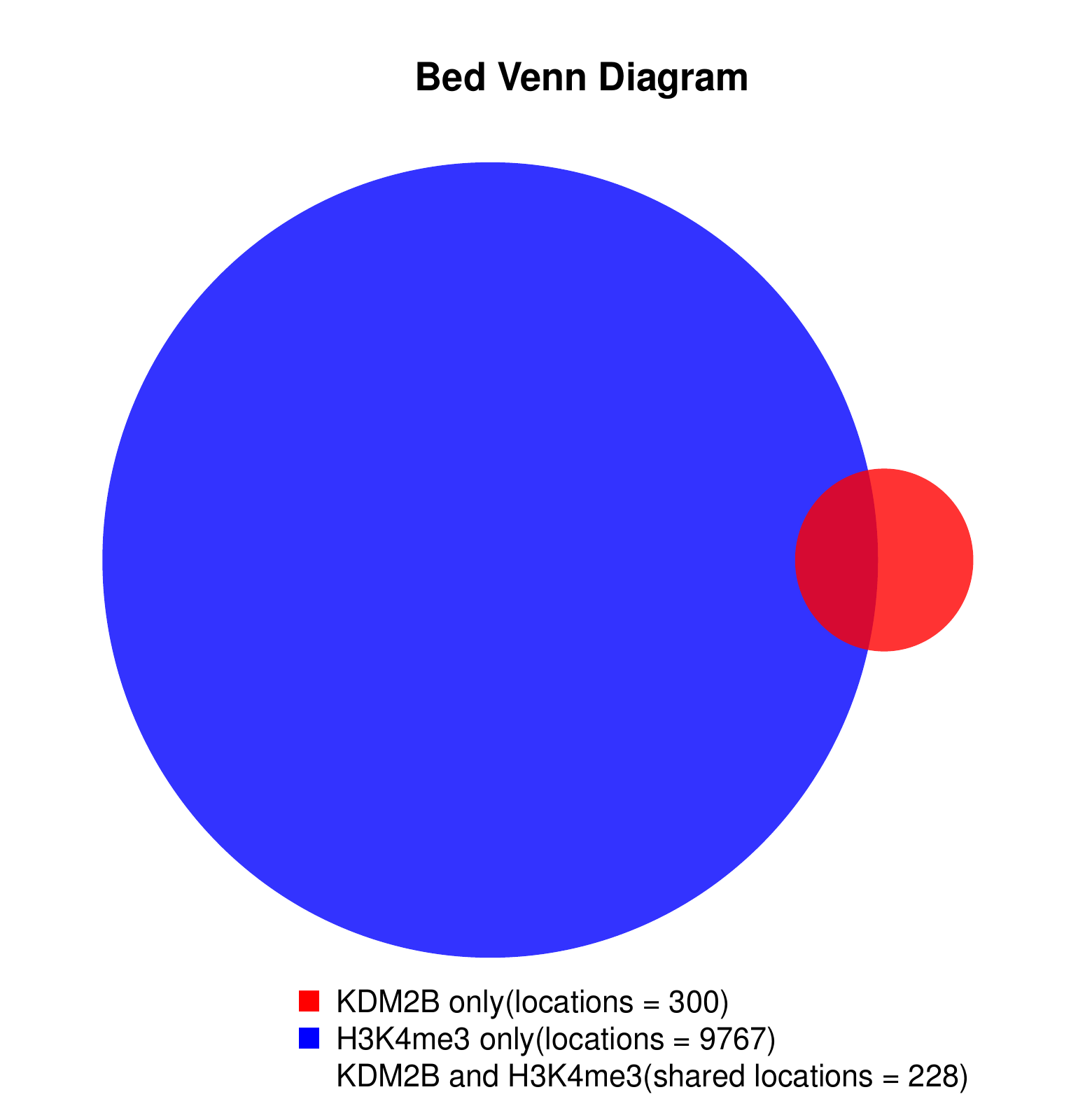
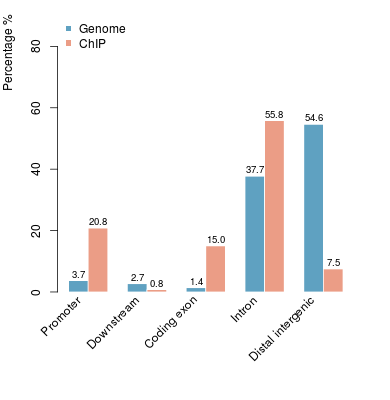
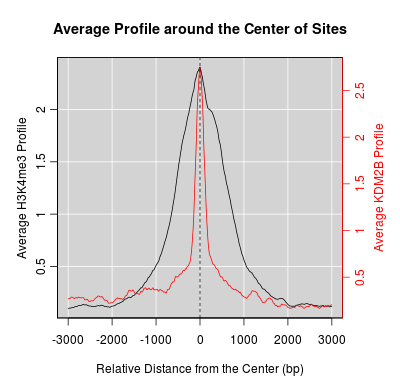
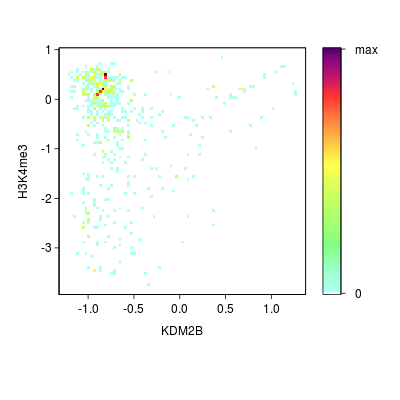
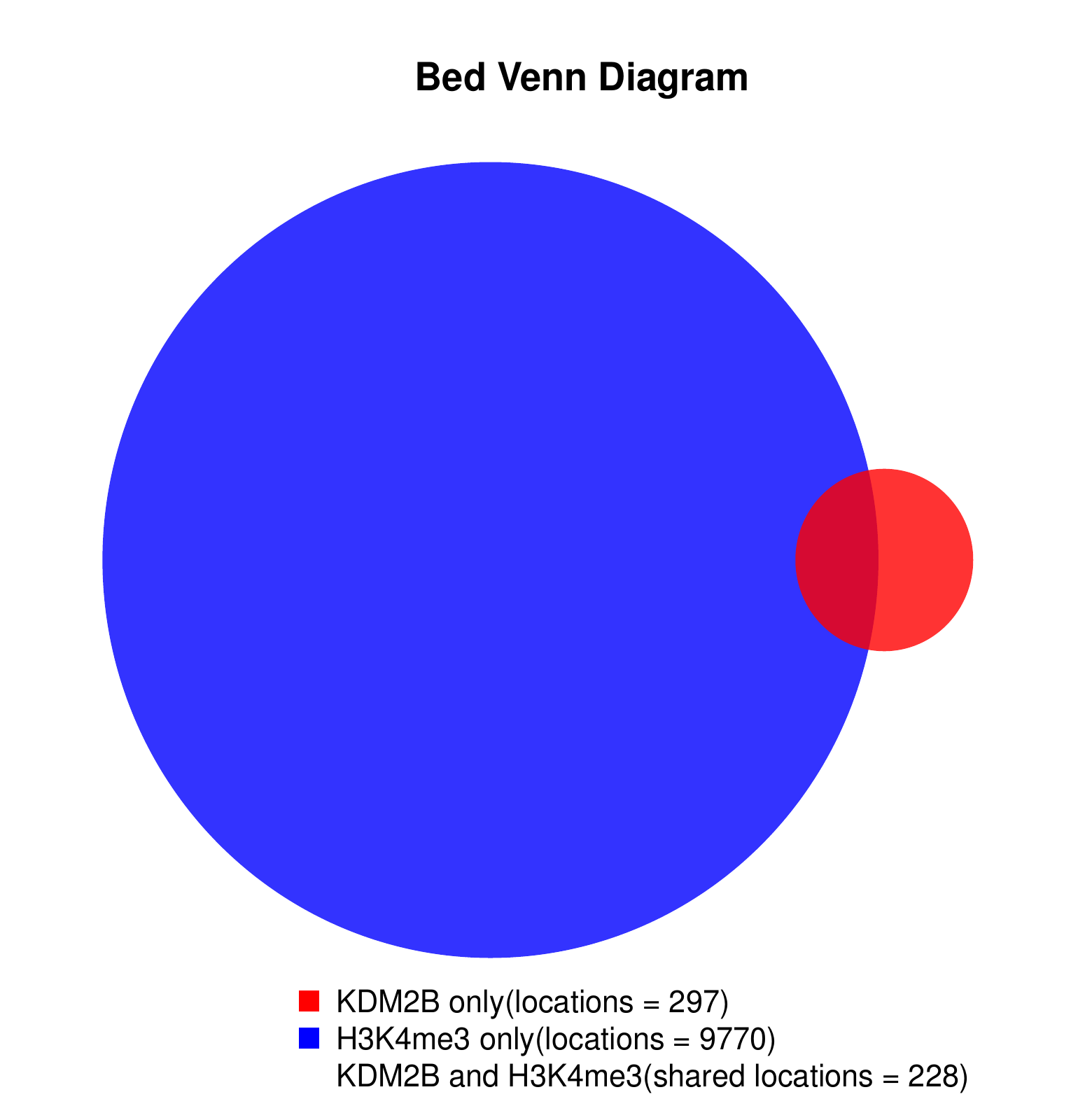
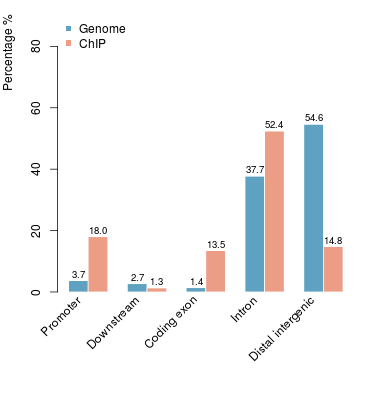
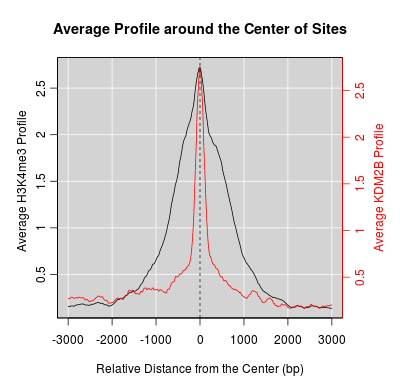
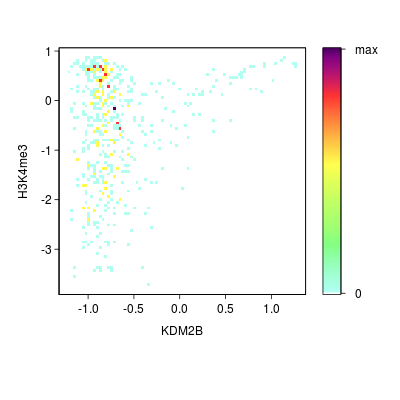
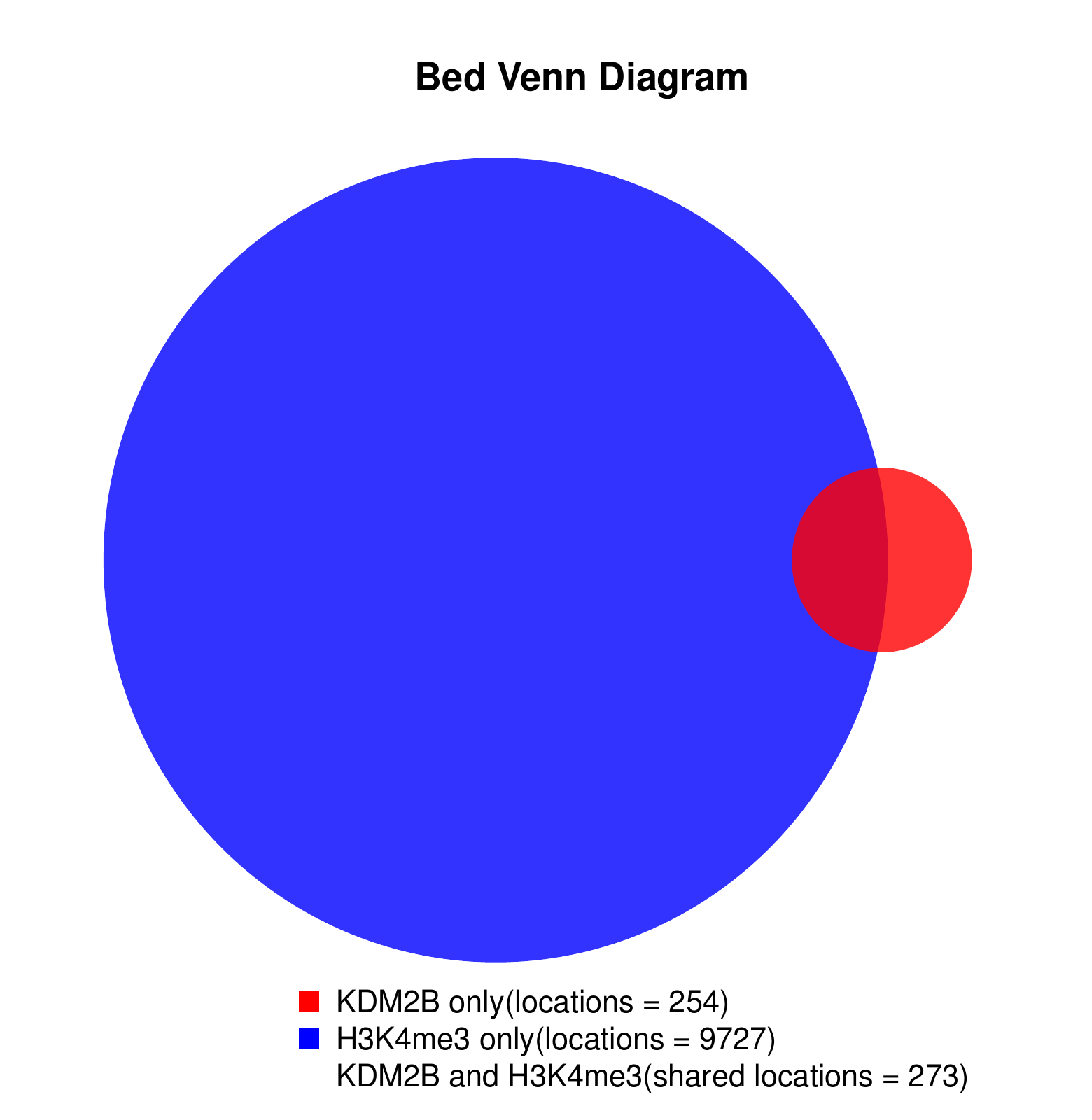
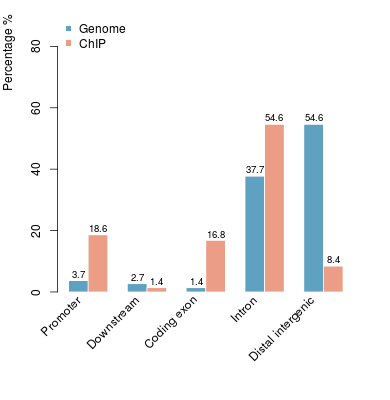
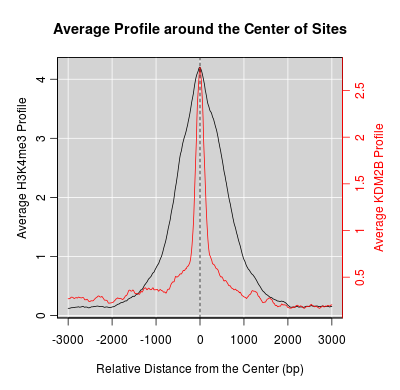
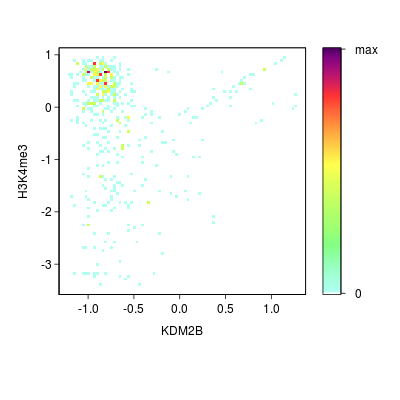
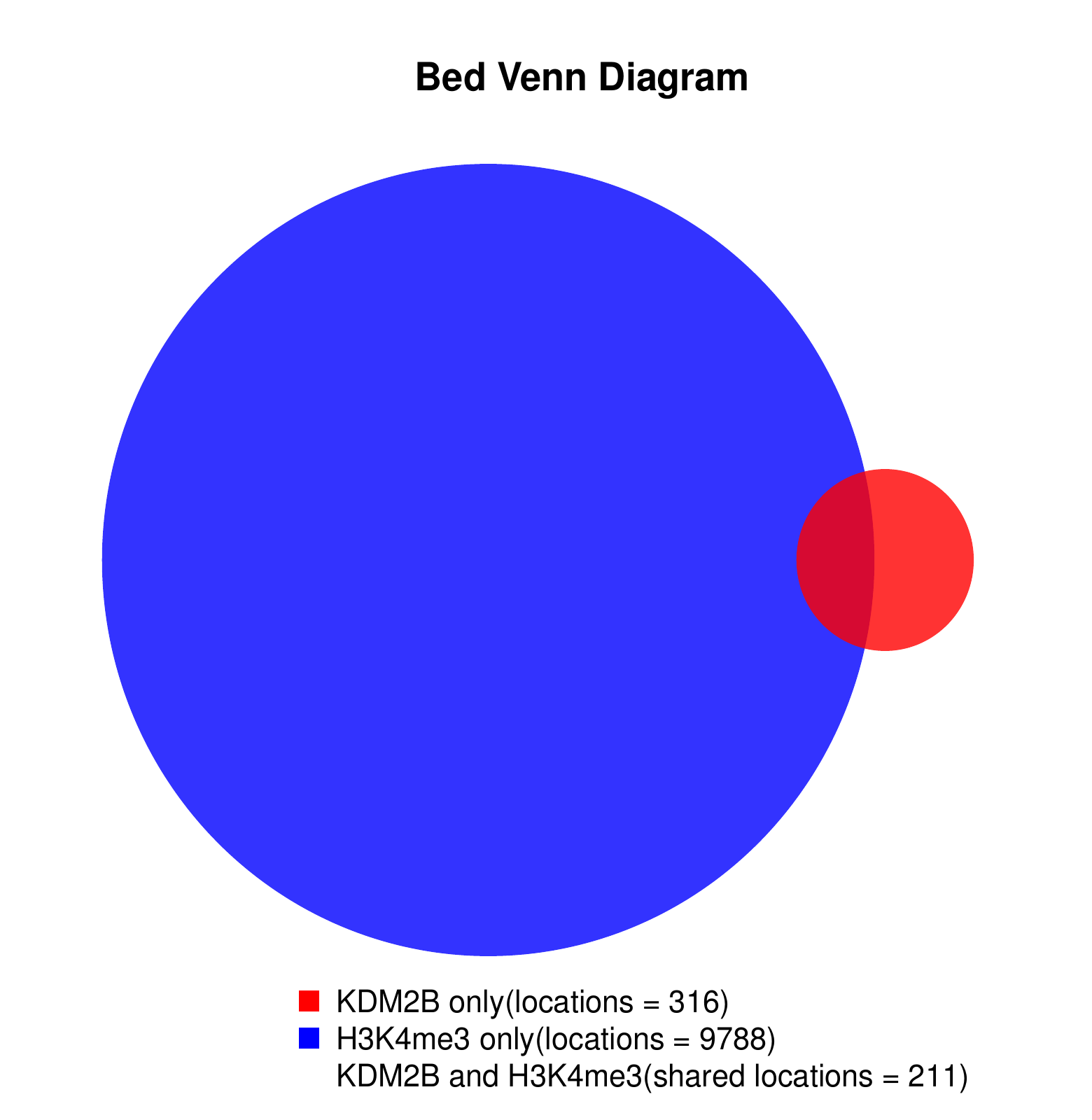
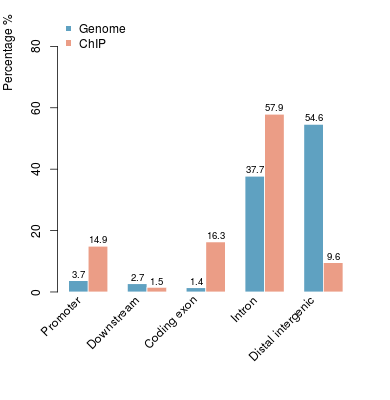
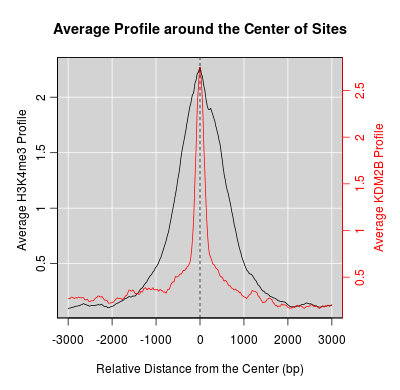
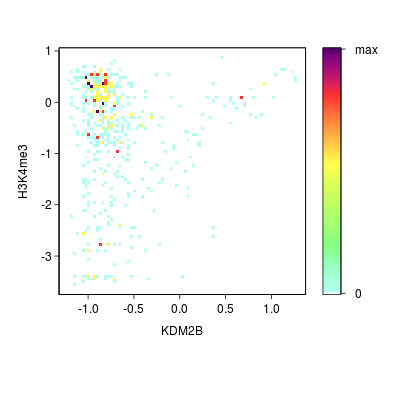
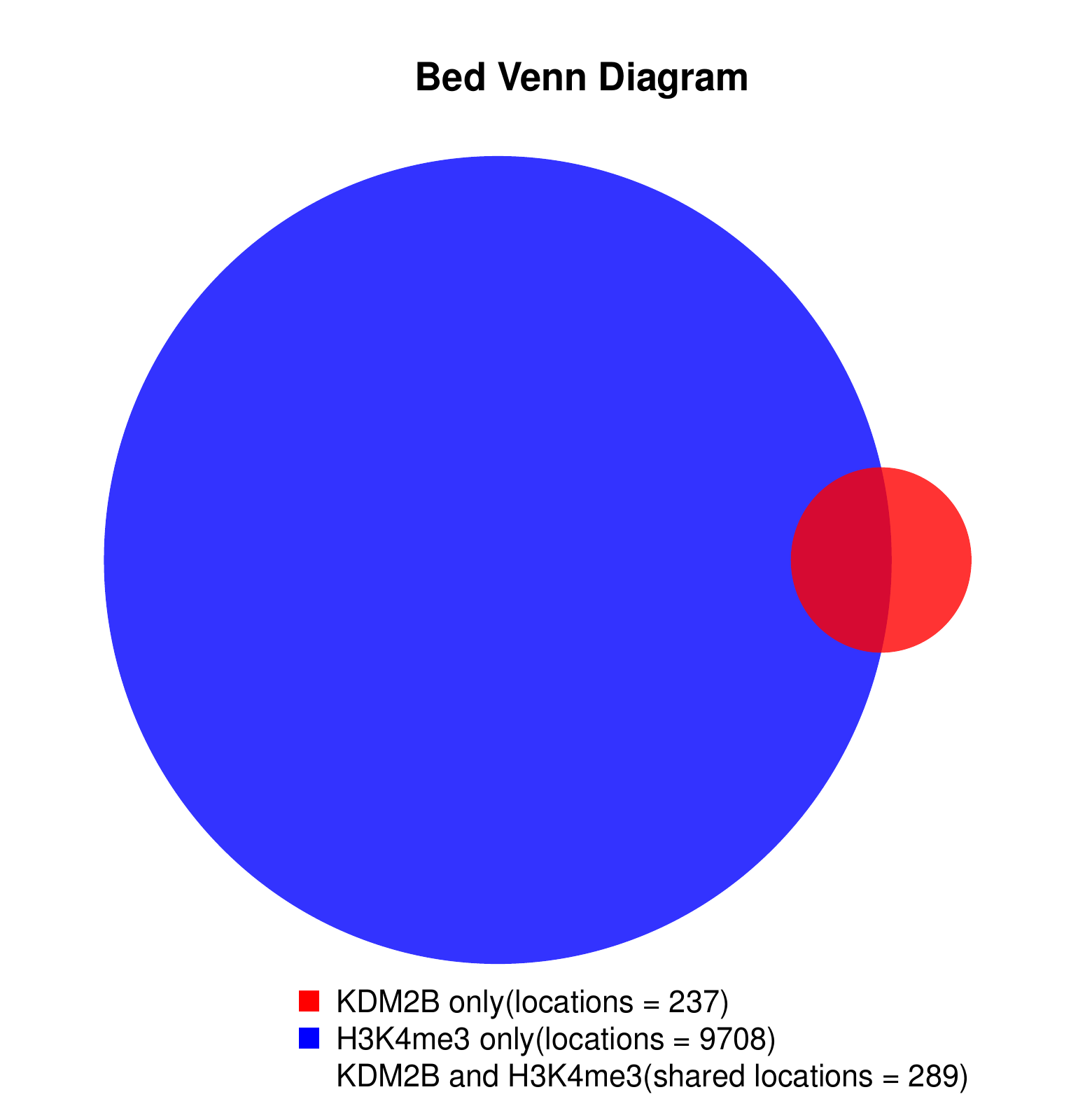
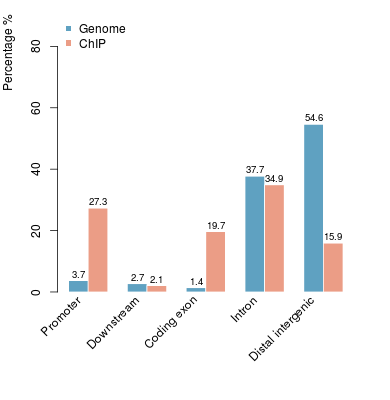
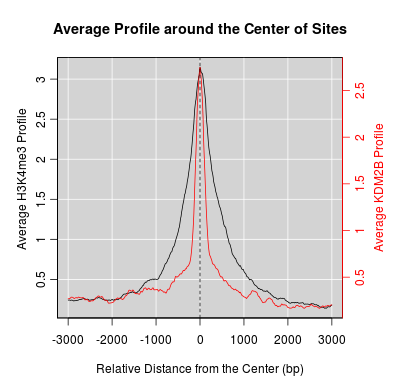
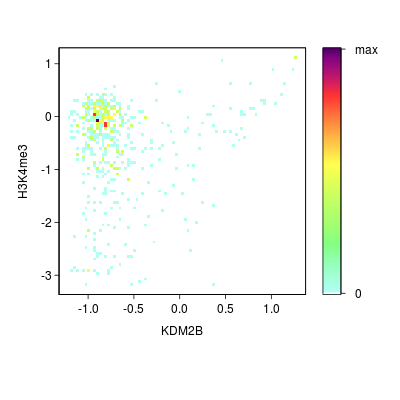
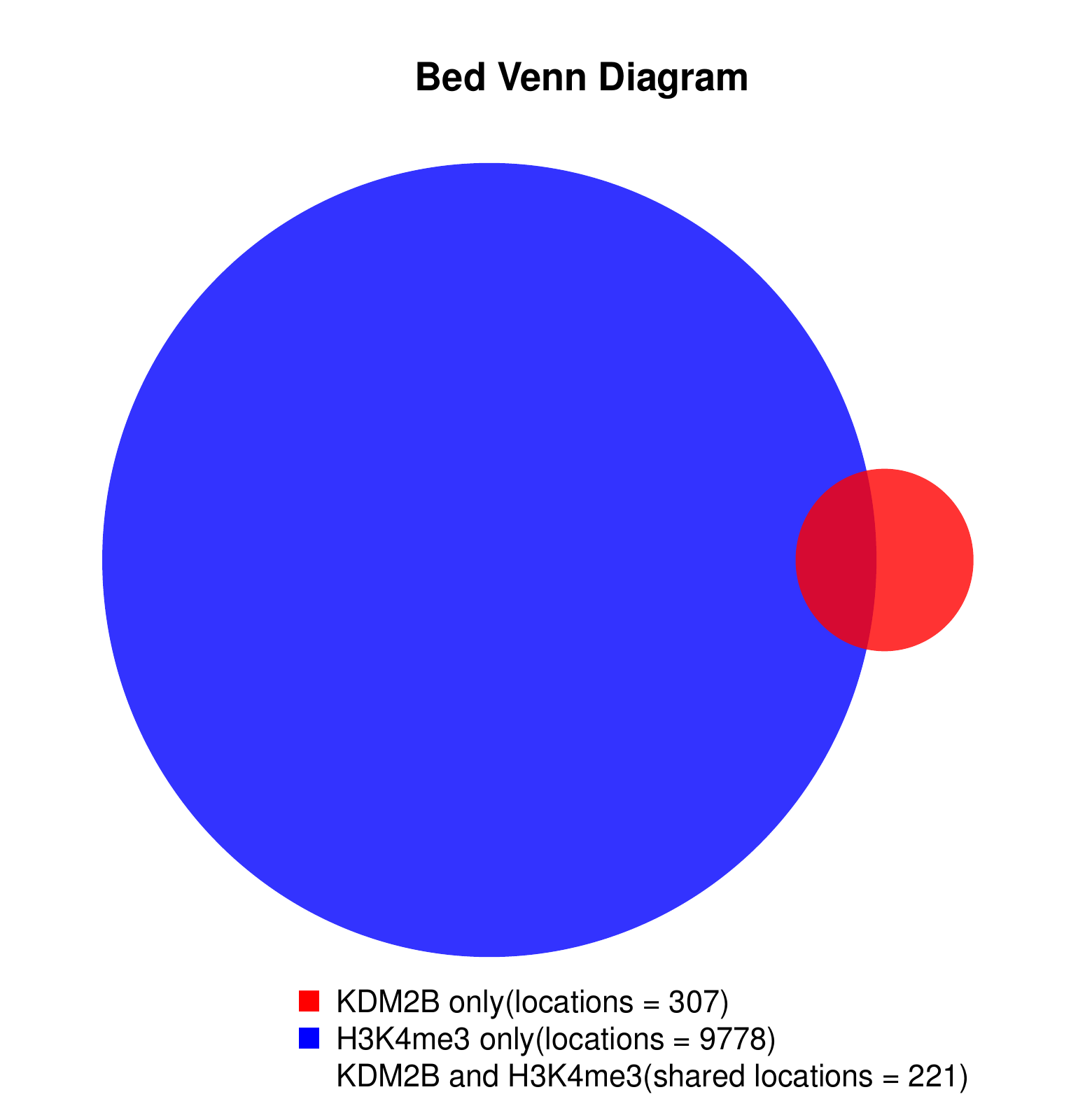
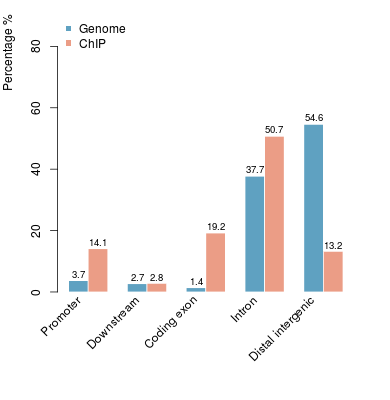
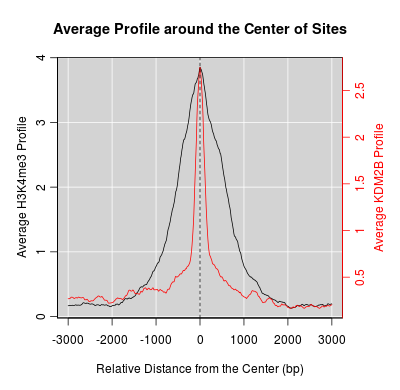
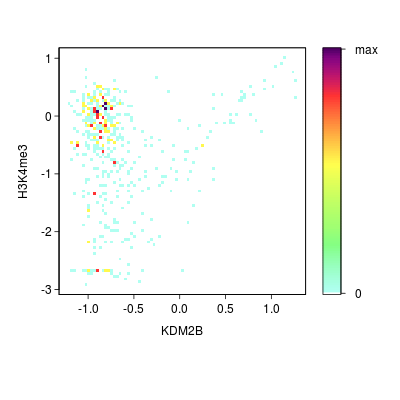
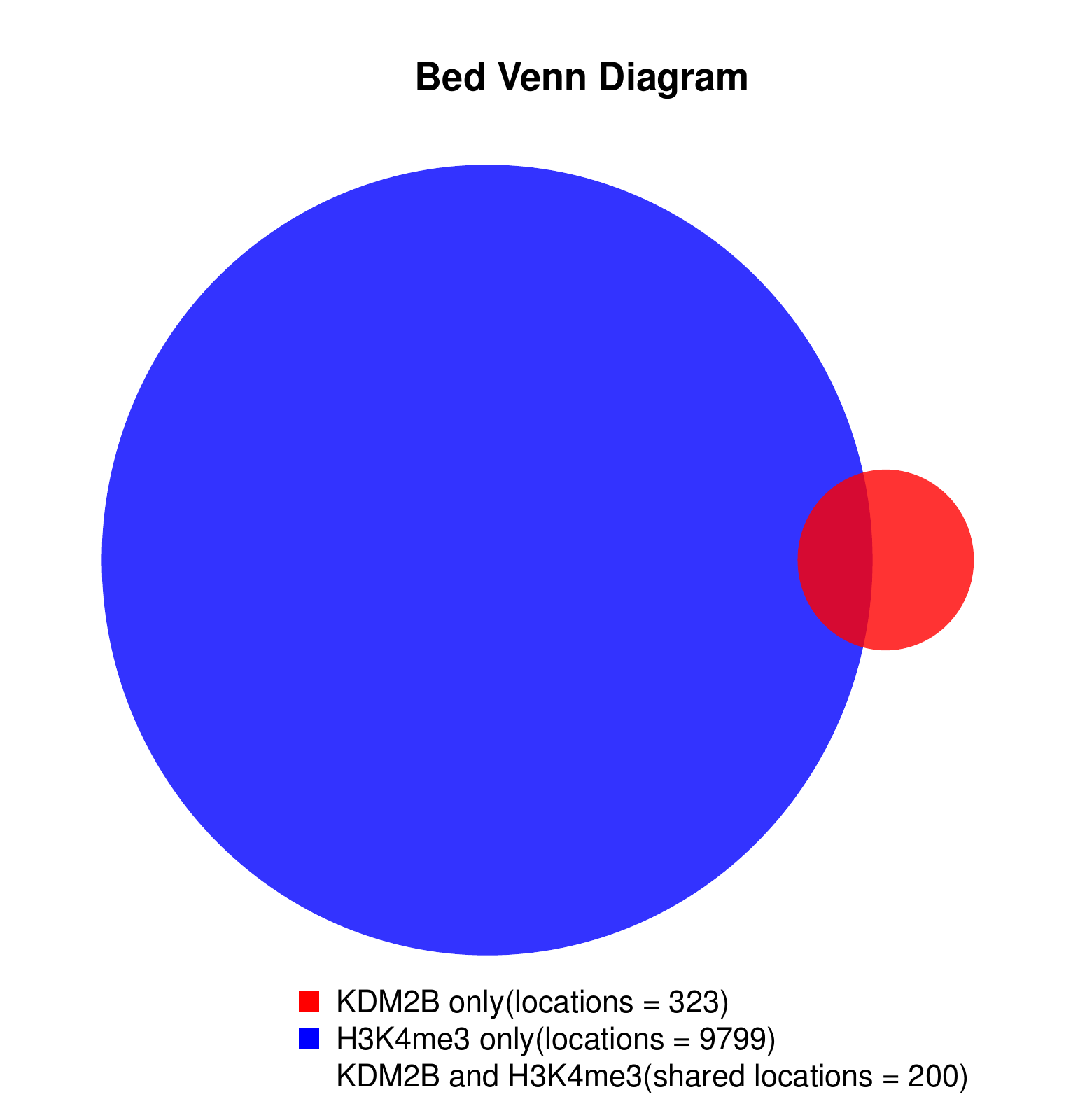
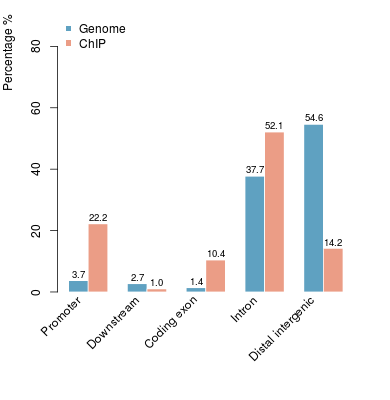
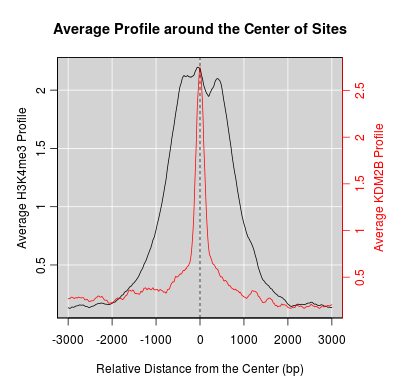
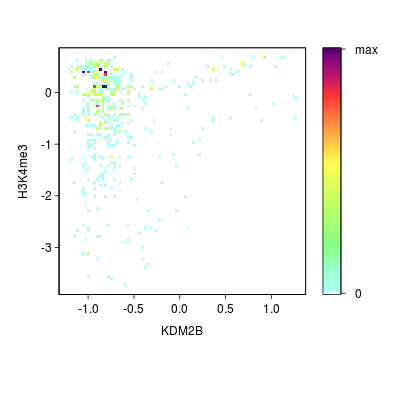
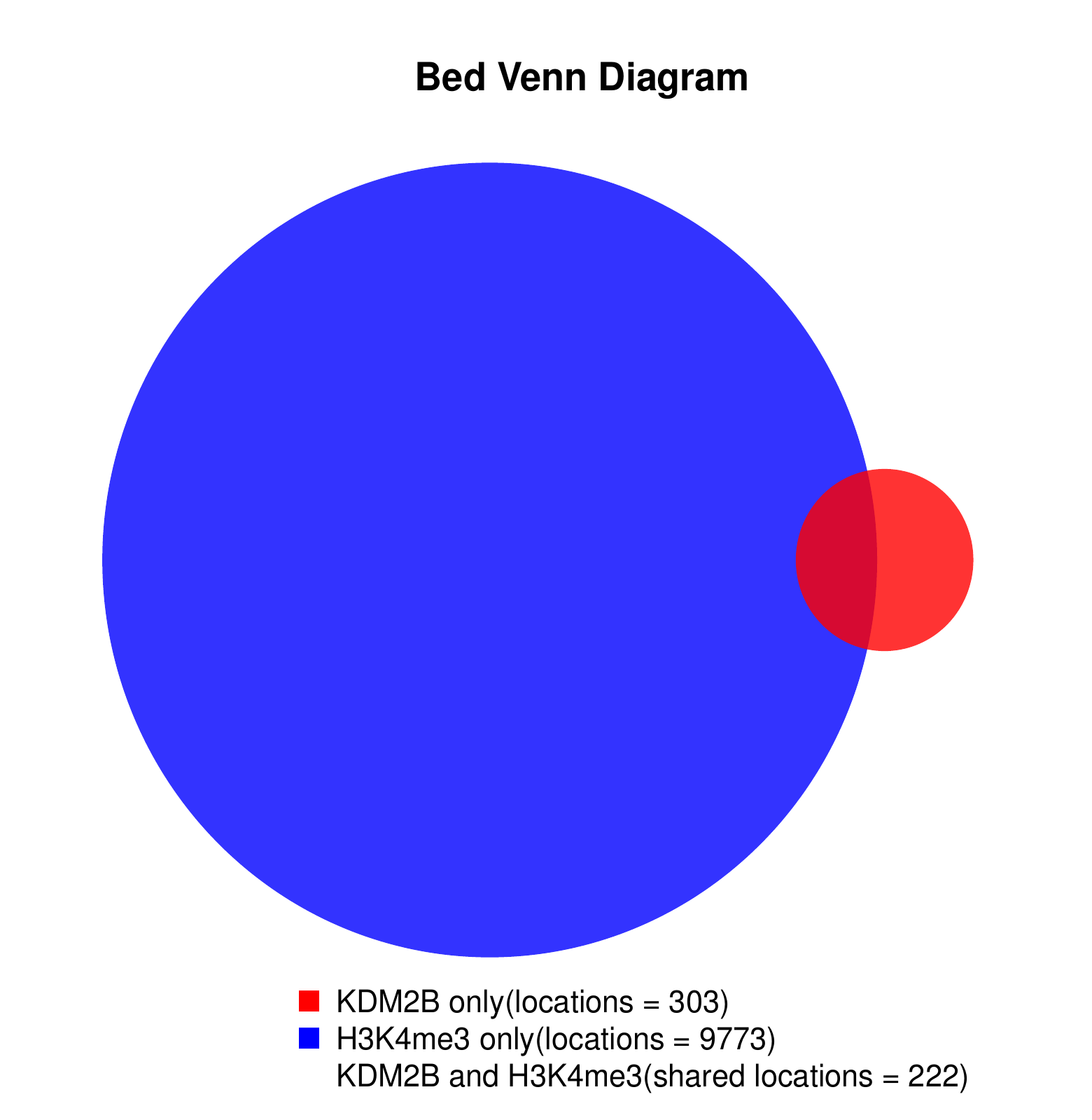
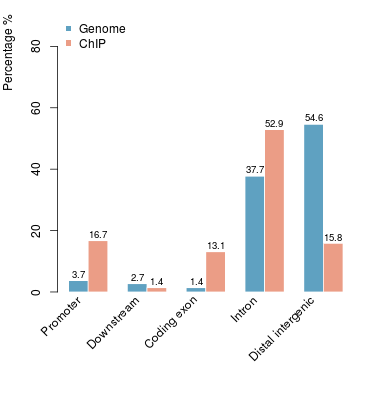
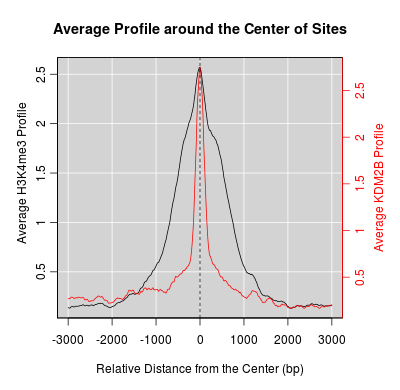
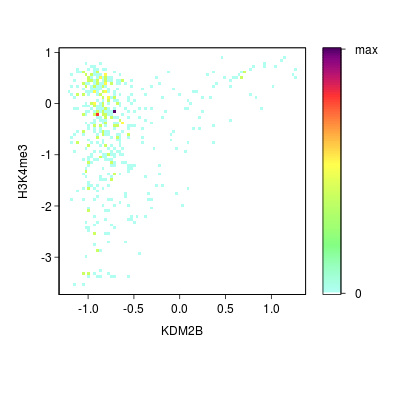
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||
KDM2B compare with H3K4me3
| ||||||||
| ||||||||