ncHMR detector
Demo dataset
Here, we provide a demo to help users to utilize the webserver. We can click mESC_GSM1562337_CBX7.bed or click example file in Home page to download CBX7's ChIP-seq peaks in mESC as a sample dataset.
Required arguments
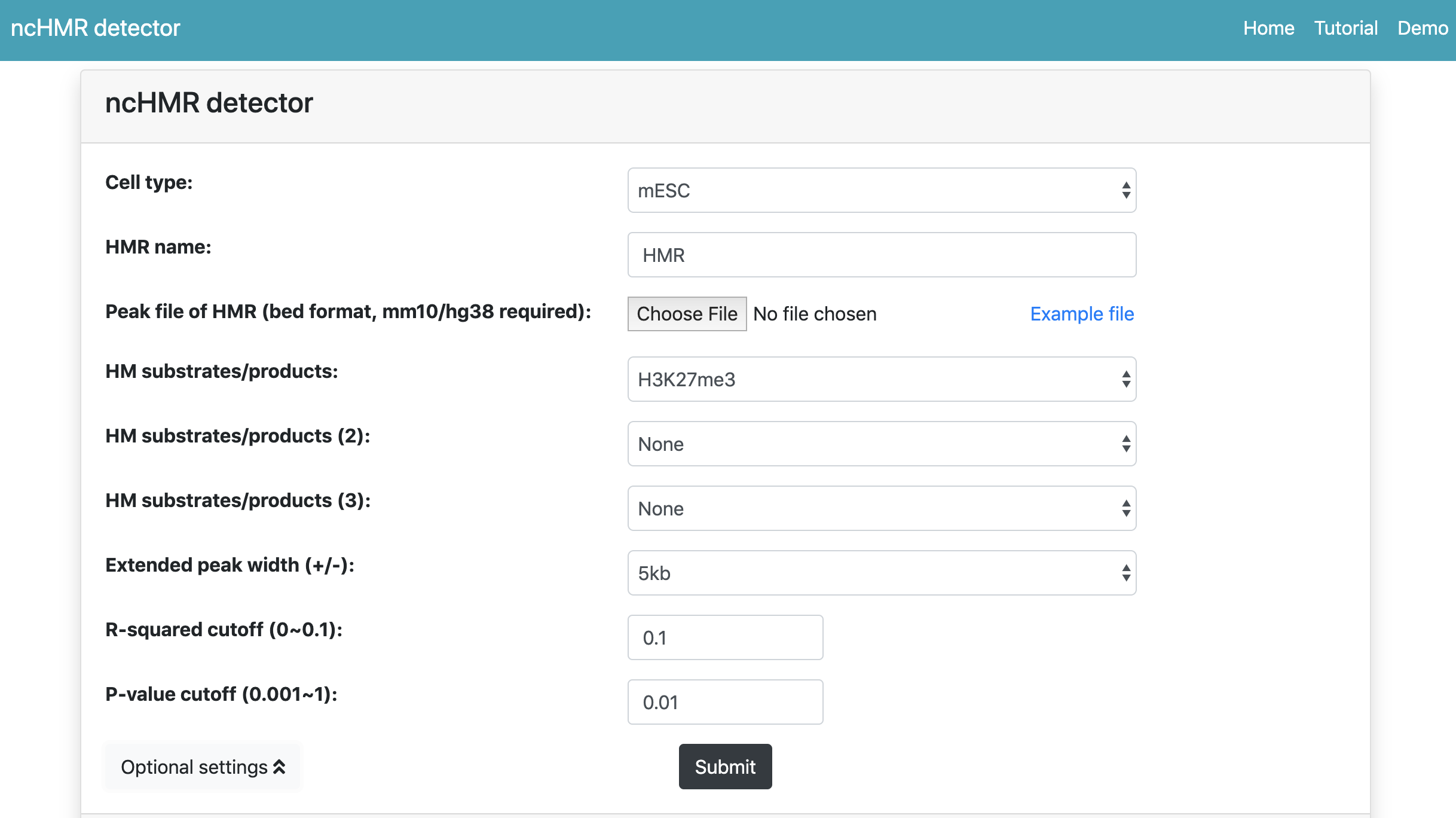
We click the Choose File button to upload sample dataset and then set several arguments as follows:
- Cell type: mESC
- HMR name: CBX7
- HM substrates/products: H3K27me3
Optional arguments
Then we can click Optinal settings to expand the form. Since H3K27me3, CBX7's classical HM substrates is a well-known broad histone modification, so we choose 5 kb as Extended peak width. All arguments are showed in Figure 1.
Output
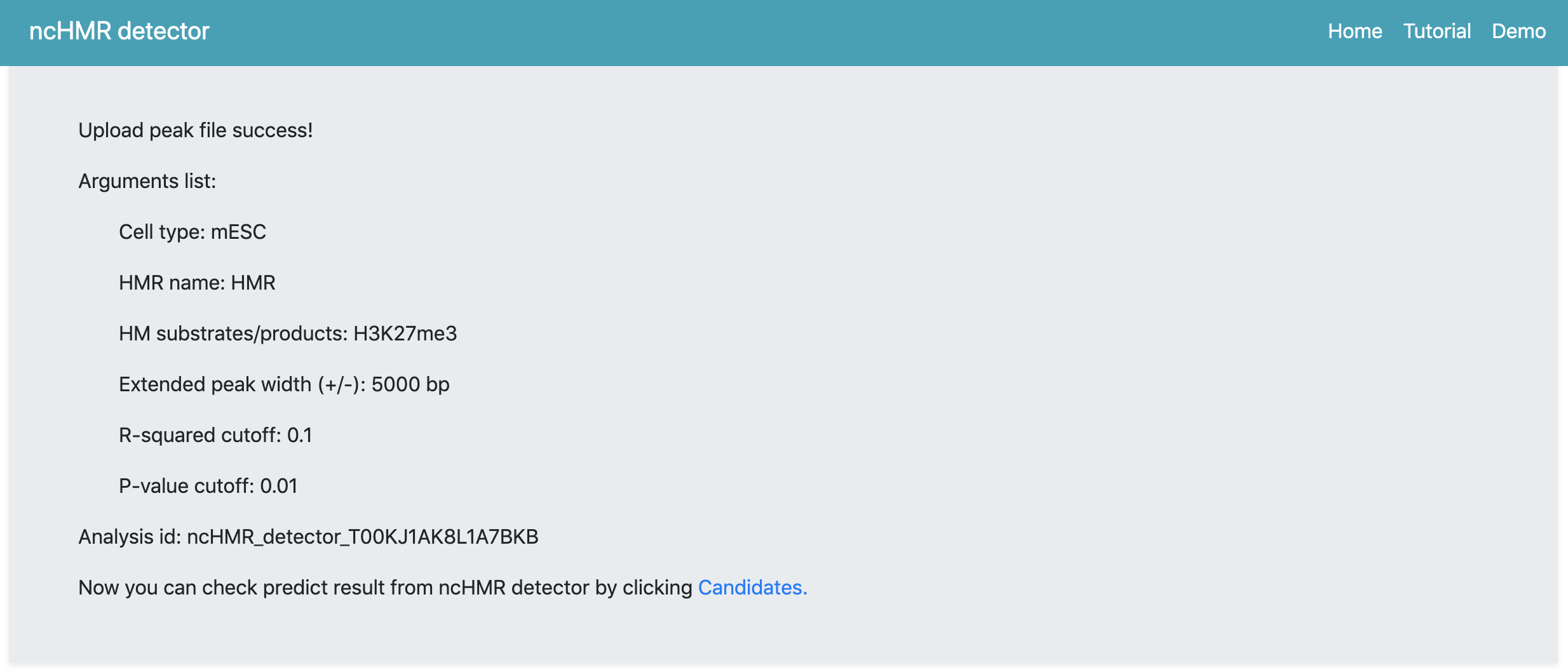
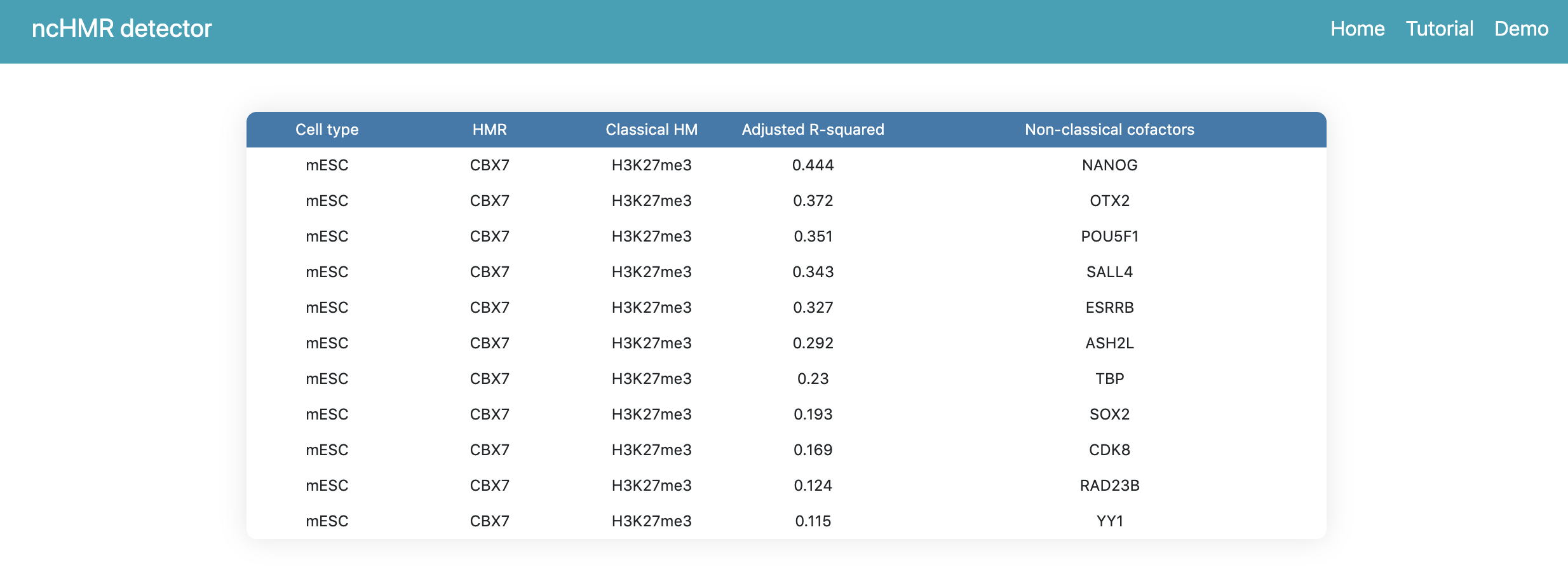
After click Submit, the webserver will pop up a window to prompt users that the analysis will begin automatically once the popup window is closed. Users are not allowed to refresh or leave the webpage during the process. After several minutes, the webserver will return a summary webpage (Figure 2). Here, we obtain a random analysis id. Then we can click Candidates buttom in the summary page to check the prediction result(Figure 3). Here, ncHMR detector detects 11 non-classical function cofactors of CBX7 with 0.1 as R-squared cutoff.